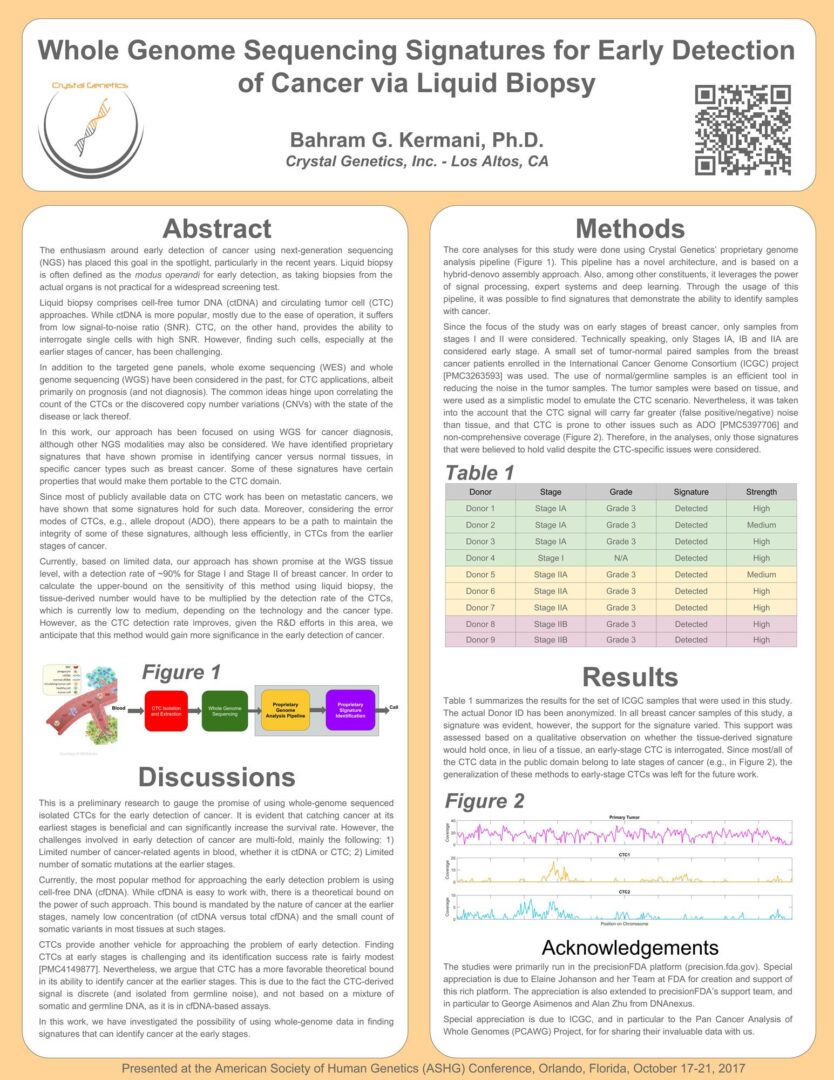
Enabling high-quality early detection of multiple cancers via liquid biopsy, CTCs and custom-designed genome analysis pipeline
CTC (Liquid Biopsy)
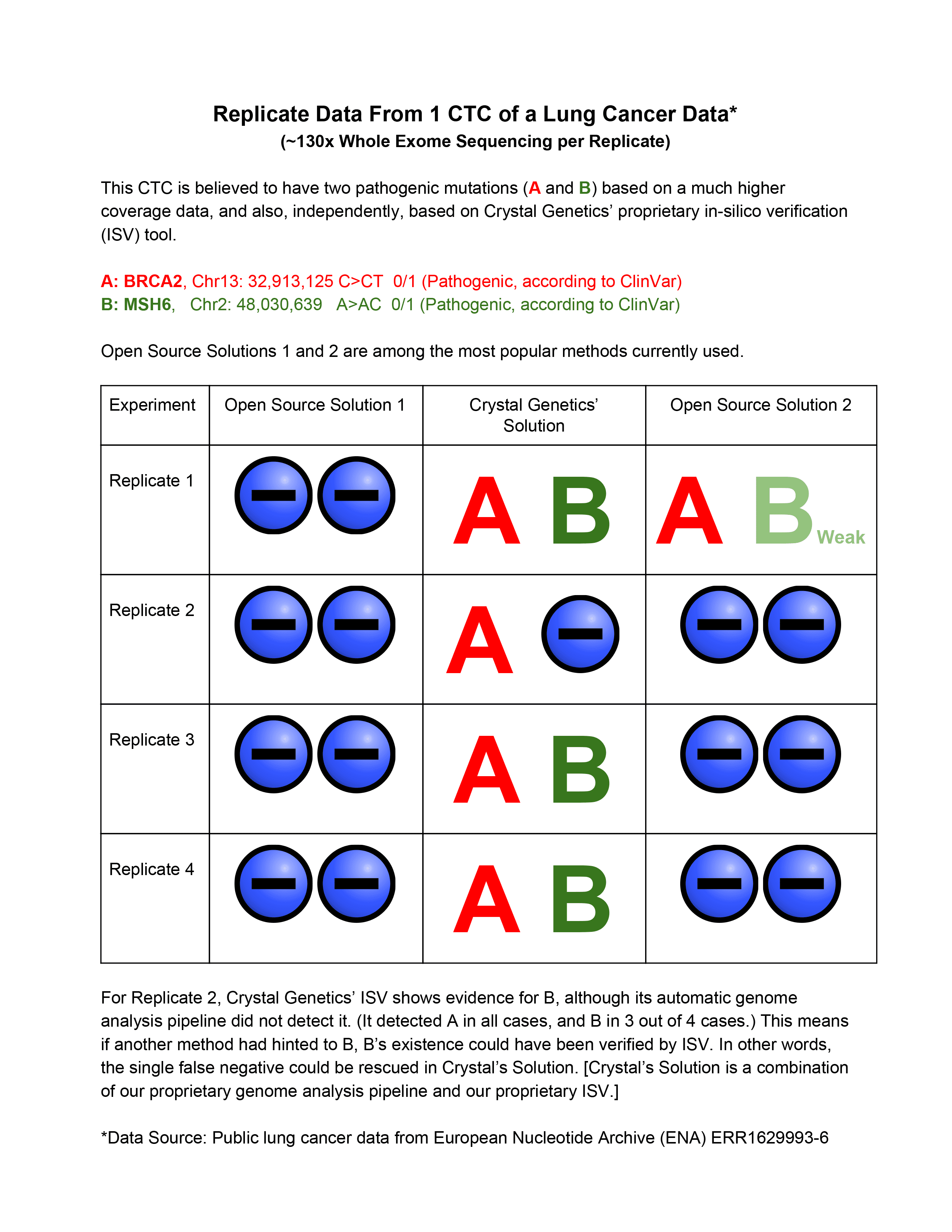
Circulating Tumor Cells (CTCs) have been successfully used in the past for prognostic applications [1]. Most of these applications rely on enumeration of CTCs or copy number variation analysis. Recently, CTCs have gained interest for mutation analysis, in different areas including diagnostic applications [2]. One such application is early detection of cancer [3,4]. Many of these applications rely on next-generation sequencing (NGS) following the CTC extraction. The analysis of CTC-derived NGS data is far from trivial. In fact, it is well known that the CTC data is subject to Allelic Dropout (ADO), false negatives and false positives. Depending on the amplification methods used, some are more prone to others. Hence, the quality of CTC-based data (especially for a small number of cells such as single cell) could be significantly compromised, as compared to the regular/bulk genome sequencing. Therefore, the bioinformatics analysis of CTC data plays a major role in the success of the underlying application. This work gives an example of the sensitivity challenge of CTC mutation discovery, and the strength of Crystal Genetics’ Proprietary genome analysis pipeline, in this regard.


CTC (Liquid Biopsy)
Circulating Tumor Cells (CTCs) have been successfully used in the past for prognostic applications [1]. Most of these applications rely on enumeration of CTCs or copy number variation analysis. Recently, CTCs have gained interest for mutation analysis, in different areas including diagnostic applications [2]. One such application is early detection of cancer [3,4]. Many of these applications rely on next-generation sequencing (NGS) following the CTC extraction. The analysis of CTC-derived NGS data is far from trivial. In fact, it is well known that the CTC data is subject to Allelic Dropout (ADO), false negatives and false positives. Depending on the amplification methods used, some are more prone to others. Hence, the quality of CTC-based data (especially for a small number of cells such as single cell) could be significantly compromised, as compared to the regular/bulk genome sequencing. Therefore, the bioinformatics analysis of CTC data plays a major role in the success of the underlying application. This work gives an example of the sensitivity challenge of CTC mutation discovery, and the strength of Crystal Genetics’ Proprietary genome analysis pipeline, in this regard.

cfDNA (Liquid Biopsy)
While the specificity of cfDNA can be increased using various filtering methods, its sensitivity remains of primary concern, mostly as a consequence of its short insert size (with the mode of ~170 bp) which makes mappability more challenging. This phenomenon cannot be remedied by increasing the coverage redundancy. Therefore the efficiency of the genome analysis method in cfDNA becomes more crucial in comparison to genomic DNA, even after discounting the effect of PCR amplification.
The raw data for the study shown here is from a 100x WGS study from an unaffected/normal person (labeled IH01), by Shendure et al. at University of Washington [1]. The FASTQ files for this sample were obtained from Illumina HiSeq2000 with 100+100 paired-end reads, and have been acquired from European Nucleotide Archive (ENA) [2]. The comparisons are done on two cancer genes (PTEN and TP53) between Crystal Genetics’ proprietary genome analysis pipeline and a popular pipeline (BWAmem+FreeBayes). The color assignments are done using Crystal Genetics’ in-silico verification (ISV) tool.
Tumor (Tissue)
The following tumor-derived genomes (WGS, FASTQ files) were obtained from Illumina’s BaseSpace. The HCC1187_Tumor data corresponds to the cell line from stage IIA, grade 3 primary ductal carcinoma (breast cancer) in a 41 year old Caucasian female. The HCC2218_Tumor data corresponds to the cell line from stage IIIA, grade 3 primary ductal carcinoma (breast cancer) in a 38 year old Caucasian female. These genomes were interrogated on all bases of the following set of 25 genes, and only the variants in the exonic regions were compared and reported. The comparisons were done between a leading pipeline (BWAmem+FreeBayes) and Crystal Genetics’ pipeline. For FreeBayes, the variants with Quality less than or equal to 250 were declared as no-calls. The discrepancies were investigated using visual/manual verification of the called variants as viewed by their raw-read support.
Gene List: APC, ATM, BARD1, BMPR1A, BRCA1, BRCA2, BRIP1, CDH1, CDK4, CDKN2A,CHEK2, EPCAM, MLH1, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD51C, RAD51D, SMAD4, STK11, TP53.

Tumor (Tissue)
The following tumor-derived genomes (WGS, FASTQ files) were obtained from Illumina’s BaseSpace. The HCC1187_Tumor data corresponds to the cell line from stage IIA, grade 3 primary ductal carcinoma (breast cancer) in a 41 year old Caucasian female. The HCC2218_Tumor data corresponds to the cell line from stage IIIA, grade 3 primary ductal carcinoma (breast cancer) in a 38 year old Caucasian female. These genomes were interrogated on all bases of the following set of 25 genes, and only the variants in the exonic regions were compared and reported. The comparisons were done between a leading pipeline (BWAmem+FreeBayes) and Crystal Genetics’ pipeline. For FreeBayes, the variants with Quality less than or equal to 250 were declared as no-calls. The discrepancies were investigated using visual/manual verification of the called variants as viewed by their raw-read support.
Gene List: APC, ATM, BARD1, BMPR1A, BRCA1, BRCA2, BRIP1, CDH1, CDK4, CDKN2A,CHEK2, EPCAM, MLH1, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD51C, RAD51D, SMAD4, STK11, TP53.
Germline
NA12878 is one of the most widely studied genomes in the world. Several luminary centers (Government and Industry) have provided variant calls for this genome.
We are glad to introduce a Cancer Gene Panel for NA12878, based on a set of 25 genes known to be highly correlated with hereditary cancers of different categories. In our panel, every single base of the mentioned genes has been interrogated.
Therefore, the provided variants lend themselves to the most comprehensive interpretation for those genes. (To showcase Crystal's accuracy, we have also provided the variant calls using Illumina’s Platinum Genomes 7.1.0.)
Cancer Panel Crystal Genetics 1.0 vs. PlatinumGenomes 7.1.0.pdf
Also, previously, we had published our data for BRCA1 and BRCA2 genes for this genome, where we demonstrated our high-accuracy as compared to the 3 top world benchmarks: Illumina’s Platinum Genomes (V7.1.0 & V8.1.0) and Cancer Gene Panel Make NIST’s Genome-in-a-Bottle (GiaB) (V2.19).
Rare Diseases
This study is to demonstrate the importance of having a comprehensive and accurate variant calling for rare diseases --in this case, ATR-X Syndrome. The raw data for this genome was obtained from Illumina’s BaseSpace [1] (NA12878_X10_Rep1_S1_V2.5).
Alpha-thalassemia mental retardation syndrome (ATRX), also called alpha-thalassemia X-linked mental retardation, nondeletion type or ATR-X syndrome is caused by mutations in the ATRX gene. Females with this mutated gene have no specific signs or features, but may demonstrate skewed X chromosome inactivation. Hemizygous males tend to be moderately intellectually disabled and have physical characteristics including coarse facial features, microcephaly (small head size), hypertelorism (widely spaced eyes), a depressed nasal bridge, a tented upper lip, and an everted lower lip. Mild or moderate anemia, associated with alpha-thalassemia, is part of the condition. [2]
In this demonstration, we have used NA12878, and not an affected individual. The objective is to show the landscape of the ATRX gene, and the performance of two pipelines on its variants, for the whole gene, and not limited to the coding region.The comparison was done between Crystal Genetics' proprietary genome analysis pipeline (fastq-to-vcf) and a leading/popular pipeline (Illumina's Isaac, provided as a part of the Base Space data). For both pipelines, if a variant was not marked as PASS, it was assumed to be a No Call in the downstream application. The discrepancies were investigated using Crystal Genetics' In-Silico Verification (ISV) tool. The assigned colors reflect the support of the variants by the raw reads, as viewed by ISV.
NA12878_X10pcrfree2p5_ATRX_CrystalGenetics_vs_Isaac
[2] https://en.wikipedia.org/wiki/Alpha-thalassemia_mental_retardation_syndrome


Rare Diseases
This study is to demonstrate the importance of having a comprehensive and accurate variant calling for rare diseases --in this case, ATR-X Syndrome. The raw data for this genome was obtained from Illumina’s BaseSpace [1] (NA12878_X10_Rep1_S1_V2.5).
Alpha-thalassemia mental retardation syndrome (ATRX), also called alpha-thalassemia X-linked mental retardation, nondeletion type or ATR-X syndrome is caused by mutations in the ATRX gene. Females with this mutated gene have no specific signs or features, but may demonstrate skewed X chromosome inactivation. Hemizygous males tend to be moderately intellectually disabled and have physical characteristics including coarse facial features, microcephaly (small head size), hypertelorism (widely spaced eyes), a depressed nasal bridge, a tented upper lip, and an everted lower lip. Mild or moderate anemia, associated with alpha-thalassemia, is part of the condition. [2]
In this demonstration, we have used NA12878, and not an affected individual. The objective is to show the landscape of the ATRX gene, and the performance of two pipelines on its variants, for the whole gene, and not limited to the coding region.The comparison was done between Crystal Genetics' proprietary genome analysis pipeline (fastq-to-vcf) and a leading/popular pipeline (Illumina's Isaac, provided as a part of the Base Space data). For both pipelines, if a variant was not marked as PASS, it was assumed to be a No Call in the downstream application. The discrepancies were investigated using Crystal Genetics' In-Silico Verification (ISV) tool. The assigned colors reflect the support of the variants by the raw reads, as viewed by ISV.
NA12878_X10pcrfree2p5_ATRX_CrystalGenetics_vs_Isaac
[2] https://en.wikipedia.org/wiki/Alpha-thalassemia_mental_retardation_syndrome